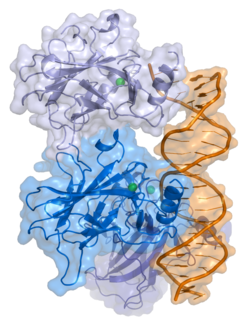
TP53
Tumorski protein P53, znan i kao p53, ćelijski tumorski-antigen p53 (UniProt ime), domaćinski gen,[5] fosfoprotein p53, tumorski supresor p53, antigen NY-CO-13 ili transformacijski vezani protein 53 (TRP53), je bilo koja proteinska izoforma proteina kodiranog homolognim genom u različitim organizmima, kao što su TP53 (ljudi) i Trp53 (miševi). Ovaj homolog (za koji se prvobitno mislilo da je, i o njemu se često govori, kao jedan protein) presudan je u višim kičmenjacima, gdje sprečava stvaranje karcinoma, te stoga funkcionira kao supresor tumora.[6] Kao takav, p53 je opisan kao "čuvar genoma" zbog svoje uloge u očuvanju stabilnosti, sprečavanjem mutacije genoma.[7] Stoga se TP53 koriste za označavanje imena gena TP53 i razlikovanje od proteina koje kodiraju (klasificiran je kao gen za supresiju tumora).[8][9][10][11][12]
Naziv 'p53' dobio je 1979. godine opisujući prividnu molekulskuu masu. Analiza SDS-PAGE pokazuje da je riječ o proteinu od 53-kilodaltona (kDa). Međutim, stvarna masa proteina p53 pune dužine (p53α) na osnovu zbroja mase aminokiselinskih ostataka iznosi samo 43,7 kDa. Ova razlika je zbog velikog broja prolinskih ostataka u proteinu, koji usporavaju njegovu migraciju na SDS-PAGE, čineći ga tako težim nego što zapravo jeste.[13] Pored proteina pune dužine, ljudski gen TP53 kodira najmanje 15 izoformnih proteina, veličine od 3,5 do 43,7 kDa. Svi ovi proteini p53 nazivaju se izoformama p53.[6] Gen TP53 je najčešće mutirani gen (> 50%) u karcinomu čovjeka, što ukazuje da TP53 ima presudnu ulogu u sprečavanju stvaranja karcinoma. Gen TP53 kodira proteine koji se vežu za DNK i regulišu ekspresiju, kako bi se spriječile mutacije genoma.[14]
Gen
urediU ljudi se gen TP53 nalazi na kratkom kraku hromosoma 17 (17p13.1).[8][9][10][11] Proteže na 20 kb, s nekodirajućim egzonom 1 i vrlo dugim intronom od 10 kb. Kodirajuća sekvenca sadrži pet regija koje imauju visok stupanj konzerviranosti kod kičmenjaka, pretežno u egzonima 2, 5, 6, 7 i 8, ali sekvence pronađene kod beskičmenjaka pokazuju samo sličnost sa sisarskim TP53.[15]Ortolozi TP53[16] su identificirani kod većine sisara za koje su dostupni potpuni podaci o genomu.
Ljudski gen TP53
urediU ljudi, uobičajeni polimorfizam uključuje zamjenu arginina za prolin na kodonskom položaju 72. Mnoge studije su istraživale genetičku vezu između ove varijacije i osjetljivost na rak; međutim, rezultati su kontroverzni. Naprimjer, meta-analiza iz 2009. godine nije pokazala vezu za rak vrata maternice.[17] Studija iz 2011. otkrila je da je mutacija prolina TP53 imala dubok utjecaj na rizik od raka gušterače kod muškaraca.[18] Studija na arapskim ženama otkrila je da je homozigotnost prolina na kodonu TP53 povezana sa smanjenim rizikom od raka dojke.[19] Jedno istraživanje sugeriralo je da polimorfizmi kodona TP53 , MDM2 SNP309 i A2164G mogu biti grupno povezani sa osjetljivošću na nerofarinksni karcinom i da je MDM2 SNP309 u kombinaciji s kodonom 72 'TP53' može ubrzati razvoj neorofarinksnog karcinoma kod žena.[20] Metaanalize iz 2011. nisu otkrile značajne veze između polimorfizama kodona 72 TP53 i oba rizika od raka debelog creva[21][22] i rizik od karcinoma endometrija.[23]
Struktura
uredip53 ima sedam domena:
- kiseli transkripcijsko-aktivacijski domen (TAD) N-kraja, poznat i kao aktivacijski domen 1 (AD1), koji aktivira faktore transkripcije. N-kraj sadrži dva komplementarna domena aktivacije transkripcije, od kojih je glavni na ostacima 1-42, a manji na ostacima 55-75, posebno uključen u regulaciju nekoliko proapoptoznih gena.[24]
- aktivacijski domen 2 (AD2) važan za apoptoznu aktivnost: ostaci 43–63.
- prolinom bogati domen važan za apoptotsku aktivnost p53 jedarnom eksportnom putem MAPK: ostaci 64–92.
- centralni dome DNK - veznog jezgra (DBD). Sadrži jedan atom cinka i nekoliko argininskih aminokiselina: ostaci 102–292. Ovo područje odgovorno je za vezivanje p53 ko-represora LMO3.[25]
- Signalizacija jedarne lokalizacije (NLS), ostaci 316–325.
- domen homooligomerizacije (OF): ostaci 307–355. Tetramerizacija je bitna za aktivnost p53 in vivo.
- C-terminal uključen u regulaciju vezanja DNK centralnog domena: ostaci 356–393.[26]
Mutacije koje deaktiviraju p53 u raku obično se javljaju u DBD. Većina ovih mutacija uništava sposobnost proteina da se veže za svoje ciljne sekvence DNK i na taj način sprečava transkripcijsku aktivaciju ovih gena. Kao takve, mutacije u DBD-u su recesivne mutacije sa gubitakom funkcije. Molekule p53 s mutacijama u OD smanjuju se s divlji tip p53 i sprečavaju ih da aktiviraju transkripciju. Stoga mutacije OD imaju dominantan negativan efekat na funkciju p53.
Divlji tip p53 je labilni protein, koji sadrži presavijene i nestrukturirane regije, koje funkcioniraju na sinergijski način.[27]
Funkcija
urediOštećenje i popravak DNK
uredip53 ima ulogu u regulaciji ili napredovanju tokom ćelijskog ciklusa, apoptoza i genomske stabilnosti pomoću nekoliko mehanizama:
- Može aktivirati popravak DNK kada DNK pretrpi oštećenje. Stoga može biti važan faktor u starenju.[28]
- Može zaustaviti rast držanjem ćelijskog ciklusana G1/S regulacijskoj tački na prepoznavanju oštećenja DNK – ako zadrži ćeliju dovoljno dugo, proteini za obnavljanje DNK će imati vrijeme da popravi štetu i ćeliji će biti omogućeno da nastavi ćelijski ciklus.
- Može pokrenuti apoptozu (tj. progrmiranu ćelijsku smrt) ako se pokaže da je oštećenje DNK nepopravljivo.
- Od suštinske je važnosti za odgovor starenja na kratke telomere .
WAF1 / CIP1 kodiranje za p21 i stotine drugih nizvodnih gena p21 (WAF1) veže se za G1 –x S / CDK (CDK4/CDK6, CDK2 i CDK1) kompleksi (važne molekule za G1 / S tranziciju u ćelijskom ciklusu) koji inhibiraju njihovu aktivnost.
Kada je p21 (WAF1) složen sa CDK2, ćelija ne može nastaviti ćelijske diobe do slijedeće faze. Mutant p53 više neće vezati DNK na učinkovit način i kao posljedica toga, protein p21 neće biti dostupan kao "zaustavni signal" za diobu ćelija.[29] Studije ljudskih embrionskih matičnih ćelija (hESC) obično opisuju nefunkcionalnu os p53-p21 G1 / S puta kontrolne tačke s naknadnim značajem za regulaciju ćelijskog ciklusa i odgovor na oštećenje DNK (DDR). Važno je da je p21 iRNK jasno prisutna i povećana regulacija nakon DDR-a u hESC-ima, ali protein p21 nije moguće otkriti. U ovom tipu ćelija, p53 aktivira brojne mikroRNK (poput miR-302a, miR-302b, miR-302c i miR-302d), koje direktno inhibiraju ekspresiju p21 u hESCs.
Protein p21 veže se direktno za ciklin-CDK komplekse, koji pokreću ćelijski ciklus i inhibira njihovu kinaznu aktivnost, uzrokujući tako zaustavljanje ćelijskog ciklusa, kako bi se omogućilo popravak. Pritom p21 također može posredovati zaustavljanje rasta povezano s diferencijacijom i trajnije zaustavljanje rasta povezano sa ćelijskom senescencijom. Gen p21 sadrži nekoliko elemenata odgovora p53 koji posreduju u direktnom vezivanju proteina p53, što rezultira transkripcijskom aktivacijom gena koji kodira protein p21.
Putevi p53 i RB1 povezani su putem p14ARF, što povećava mogućnost da se mogu međusobno regulirati.[30]
Ekspresiju p53 može podstaknuti UV-svjetlost, koja također uzrokuje oštećenje DNK. U ovom slučaju, p53 može inicirati događaje koji vode do sunčanja.[31][32]
Interakcije
urediPokazano je da p53 uspostavlja interakcije sa:
- AIMP2,[33]
- ANKRD2,[34]
- APTX,[35]
- ATM,[36][37][38][39][40]
- ATR,[36][37]
- ATF3,[41][42]
- AURKA,[43]
- BAK1,[44]
- BARD1,[45]
- BLM,[46][47][48][49]
- BRCA1,[45][50][51][52][53]
- BRCA2,[45][54]
- BRCC3,[45]
- BRE,[45]
- CEBPZ,[55]
- CDC14A,[56]
- Cdk1,[57][58]
- CFLAR,[59]
- CHEK1,[46][60][61]
- CCNG1,[62]
- CREBBP,[63][64][65]
- CREB1,[65]
- Ciklin H,[66]
- CDK7,[66][67]
- DNA-PKcs,[37][60][68]
- E4F1,[69][70]
- EFEMP2,[71]
- EIF2AK2,[72]
- ELL,[73]
- EP300,[64][74][75][76]
- ERCC6,[77][78]
- GNL3,[79]
- GPS2,[80]
- GSK3B,[81]
- HSP90AA1,[82][83][84]
- HIF1A,[85][86][87][88]
- HIPK1,[89]
- HIPK2,[90][91]
- HMGB1,[92][93]
- HSPA9,[94]
- Huntingtin,[95]
- ING1,[96][97]
- ING4,[98][99]
- ING5,[98]
- IκBα,[100]
- KPNB1,[82]
- LMO3,[25]
- Mdm2,[63][101][102][103]
- MDM4,[104][105]
- MED1,[106][107]
- MAPK9,[108][109]
- MNAT1,[67]
- NDN,[110]
- NCL,[111]
- NUMB,[112]
- NF-κB,[113]
- P16,[69][103][114]
- PARC,[115]
- PARP1,[35][116]
- PIAS1,[71][117]
- CDC14B,[56]
- PIN1,[118][119]
- PLAGL1,[120]
- PLK3,[121][122]
- PRKRA,[123]
- PHB,[124]
- PML,[101][125][126]
- PSME3,[127]
- PTEN,[102]
- PTK2,[128]
- PTTG1,[129]
- RAD51,[45][130][131]
- RCHY1,[132][133]
- RELA,[113]
- Reprimo
- RPA1,[134][135]
- RPL11,[114]
- S100B,[136]
- SUMO1,[137][138]
- SMARCA4,[139]
- SMARCB1,[139]
- SMN1,[140]
- STAT3,[113]
- TBP,[141][142]
- TFAP2A,[143]
- TFDP1,[144]
- TIGAR,[145]
- TOP1,[146][147]
- TOP2A,[148]
- TP53BP1,[46][149][150][151][152][153][154]
- TP53BP2,[154][155]
- TOP2B,[148]
- TP53INP1,[156][157]
- TSG101,[158]
- UBE2A,[159]
- UBE2I,[71][137][160][161]
- UBC,[33][127][138][162][163][164][165][166]
- USP7,[167]
- WRN,[49][168]
- WWOX,[169]
- XPB,[77]
- YBX1,[34][170]
- YPEL3,[171]
- YWHAZ,[172]
- Zif268,[173]
- ZNF148,[174]
- SIRT1,[175]
- circRNA_014511.[176]
Takođert pogledajte
uredi- Pifitrin, inhibitor za P53
Reference
uredi- ^ a b c GRCh38: Ensembl release 89: ENSG00000141510 - Ensembl, maj 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000059552 - Ensembl, maj 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Toufektchan, E.; Toledo, F. (2018). "The Guardian of the Genome Revisited: P53 Downregulates Genes Required for Telomere Maintenance, DNA Repair, and Centromere Structure". Cancers. 10 (5): 135. doi:10.3390/cancers10050135. PMC 5977108. PMID 29734785.
- ^ a b Surget S, Khoury MP, Bourdon JC (decembar 2013). "Uncovering the role of p53 splice variants in human malignancy: a clinical perspective". OncoTargets and Therapy. 7: 57–68. doi:10.2147/OTT.S53876. PMC 3872270. PMID 24379683.
- ^ Read AP, Strachan T (1999). "Chapter 18: Cancer Genetics". Human molecular genetics 2. New York: Wiley. ISBN 978-0-471-33061-5.
- ^ a b Matlashewski G, Lamb P, Pim D, Peacock J, Crawford L, Benchimol S (decembar 1984). "Isolation and characterization of a human p53 cDNA clone: expression of the human p53 gene". The EMBO Journal. 3 (13): 3257–62. doi:10.1002/j.1460-2075.1984.tb02287.x. PMC 557846. PMID 6396087.
- ^ a b Isobe M, Emanuel BS, Givol D, Oren M, Croce CM (1986). "Localization of gene for human p53 tumour antigen to band 17p13". Nature. 320 (6057): 84–5. Bibcode:1986Natur.320...84I. doi:10.1038/320084a0. PMID 3456488. S2CID 4310476.
- ^ a b Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B (juni 1991). "Identification of p53 as a sequence-specific DNA-binding protein". Science. 252 (5013): 1708–11. Bibcode:1991Sci...252.1708K. doi:10.1126/science.2047879. PMID 2047879. S2CID 19647885.
- ^ a b McBride OW, Merry D, Givol D (januar 1986). "The gene for human p53 cellular tumor antigen is located on chromosome 17 short arm (17p13)". Proceedings of the National Academy of Sciences of the United States of America. 83 (1): 130–4. Bibcode:1986PNAS...83..130M. doi:10.1073/pnas.83.1.130. PMC 322805. PMID 3001719.
- ^ Greška kod citiranja: Nevaljana oznaka
<ref>; nije naveden tekst za reference s imenomBourdon - ^ Ziemer MA, Mason A, Carlson DM (septembar 1982). "Cell-free translations of proline-rich protein mRNAs". The Journal of Biological Chemistry. 257 (18): 11176–80. doi:10.1016/S0021-9258(18)33948-6. PMID 7107651.
- ^ Levine AJ, Lane DP, ured. (2010). The p53 family. Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. ISBN 978-0-87969-830-0.
- ^ May P, May E (decembar 1999). "Twenty years of p53 research: structural and functional aspects of the p53 protein". Oncogene. 18 (53): 7621–36. doi:10.1038/sj.onc.1203285. PMID 10618702.
- ^ "OrthoMaM phylogenetic marker: TP53 coding sequence". Arhivirano s originala, 17. 3. 2018. Pristupljeno 2. 12. 2009.
- ^ Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, Brenna SM, et al. (august 2009). "TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies". The Lancet. Oncology. 10 (8): 772–84. doi:10.1016/S1470-2045(09)70187-1. PMID 19625214.
- ^ Sonoyama T, Sakai A, Mita Y, Yasuda Y, Kawamoto H, Yagi T, Yoshioka M, Mimura T, Nakachi K, Ouchida M, Yamamoto K, Shimizu K (2011). "TP53 codon 72 polymorphism is associated with pancreatic cancer risk in males, smokers and drinkers". Molecular Medicine Reports. 4 (3): 489–95. doi:10.3892/mmr.2011.449. PMID 21468597.
- ^ Alawadi S, Ghabreau L, Alsaleh M, Abdulaziz Z, Rafeek M, Akil N, Alkhalaf M (septembar 2011). "P53 gene polymorphisms and breast cancer risk in Arab women". Medical Oncology. 28 (3): 709–15. doi:10.1007/s12032-010-9505-4. PMID 20443084. S2CID 207372095.
- ^ Yu H, Huang YJ, Liu Z, Wang LE, Li G, Sturgis EM, Johnson DG, Wei Q (septembar 2011). "Effects of MDM2 promoter polymorphisms and p53 codon 72 polymorphism on risk and age at onset of squamous cell carcinoma of the head and neck". Molecular Carcinogenesis. 50 (9): 697–706. doi:10.1002/mc.20806. PMC 3142329. PMID 21656578.
- ^ Piao JM, Kim HN, Song HR, Kweon SS, Choi JS, Yun WJ, Kim YC, Oh IJ, Kim KS, Shin MH (septembar 2011). "p53 codon 72 polymorphism and the risk of lung cancer in a Korean population". Lung Cancer. 73 (3): 264–7. doi:10.1016/j.lungcan.2010.12.017. PMID 21316118.
- ^ Wang JJ, Zheng Y, Sun L, Wang L, Yu PB, Dong JH, Zhang L, Xu J, Shi W, Ren YC (novembar 2011). "TP53 codon 72 polymorphism and colorectal cancer susceptibility: a meta-analysis". Molecular Biology Reports. 38 (8): 4847–53. doi:10.1007/s11033-010-0619-8. PMID 21140221. S2CID 11730631.
- ^ Jiang DK, Yao L, Ren WH, Wang WZ, Peng B, Yu L (decembar 2011). "TP53 Arg72Pro polymorphism and endometrial cancer risk: a meta-analysis". Medical Oncology. 28 (4): 1129–35. doi:10.1007/s12032-010-9597-x. PMID 20552298. S2CID 32990396.
- ^ Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L (august 1998). "The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression". The EMBO Journal. 17 (16): 4668–79. doi:10.1093/emboj/17.16.4668. PMC 1170796. PMID 9707426.
- ^ a b Larsen S, Yokochi T, Isogai E, Nakamura Y, Ozaki T, Nakagawara A (februar 2010). "LMO3 interacts with p53 and inhibits its transcriptional activity". Biochemical and Biophysical Research Communications. 392 (3): 252–7. doi:10.1016/j.bbrc.2009.12.010. PMID 19995558.
- ^ Harms KL, Chen X (mart 2005). "The C terminus of p53 family proteins is a cell fate determinant". Molecular and Cellular Biology. 25 (5): 2014–30. doi:10.1128/MCB.25.5.2014-2030.2005. PMC 549381. PMID 15713654.
- ^ Bell S, Klein C, Müller L, Hansen S, Buchner J (oktobar 2002). "p53 contains large unstructured regions in its native state". Journal of Molecular Biology. 322 (5): 917–27. doi:10.1016/S0022-2836(02)00848-3. PMID 12367518.
- ^ Gilbert, Scott F. Developmental Biology, 10th ed. Sunderland, MA USA: Sinauer Associates, Inc. Publishers. str. 588.
- ^ National Center for Biotechnology Information (1998). The p53 tumor suppressor protein. Genes and Disease. United States National Institutes of Health. Pristupljeno 28. 5. 2008.
- ^ Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH (septembar 1998). "p14ARF links the tumour suppressors RB and p53". Nature. 395 (6698): 124–5. Bibcode:1998Natur.395..124B. doi:10.1038/25867. PMID 9744267. S2CID 4355786.
- ^ "Genome's guardian gets a tan started". New Scientist. 17. 3. 2007. Pristupljeno 29. 3. 2007.
- ^ Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter SR, Fisher DE (mart 2007). "Central role of p53 in the suntan response and pathologic hyperpigmentation". Cell. 128 (5): 853–64. doi:10.1016/j.cell.2006.12.045. PMID 17350573.
- ^ a b Han JM, Park BJ, Park SG, Oh YS, Choi SJ, Lee SW, Hwang SK, Chang SH, Cho MH, Kim S (august 2008). "AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53". Proceedings of the National Academy of Sciences of the United States of America. 105 (32): 11206–11. Bibcode:2008PNAS..10511206H. doi:10.1073/pnas.0800297105. PMC 2516205. PMID 18695251.
- ^ a b Kojic S, Medeot E, Guccione E, Krmac H, Zara I, Martinelli V, Valle G, Faulkner G (maj 2004). "The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle". Journal of Molecular Biology. 339 (2): 313–25. doi:10.1016/j.jmb.2004.03.071. PMID 15136035.
- ^ a b Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, Lavin MF (maj 2004). "Aprataxin, a novel protein that protects against genotoxic stress". Human Molecular Genetics. 13 (10): 1081–93. doi:10.1093/hmg/ddh122. PMID 15044383.
- ^ a b Fabbro M, Savage K, Hobson K, Deans AJ, Powell SN, McArthur GA, Khanna KK (juli 2004). "BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage". The Journal of Biological Chemistry. 279 (30): 31251–8. doi:10.1074/jbc.M405372200. PMID 15159397.
- ^ a b c Kim ST, Lim DS, Canman CE, Kastan MB (decembar 1999). "Substrate specificities and identification of putative substrates of ATM kinase family members". The Journal of Biological Chemistry. 274 (53): 37538–43. doi:10.1074/jbc.274.53.37538. PMID 10608806.
- ^ Kang J, Ferguson D, Song H, Bassing C, Eckersdorff M, Alt FW, Xu Y (januar 2005). "Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression". Molecular and Cellular Biology. 25 (2): 661–70. doi:10.1128/MCB.25.2.661-670.2005. PMC 543410. PMID 15632067.
- ^ Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller SP, Lavin MF (decembar 1998). "ATM associates with and phosphorylates p53: mapping the region of interaction". Nature Genetics. 20 (4): 398–400. doi:10.1038/3882. PMID 9843217. S2CID 23994762.
- ^ Westphal CH, Schmaltz C, Rowan S, Elson A, Fisher DE, Leder P (maj 1997). "Genetic interactions between atm and p53 influence cellular proliferation and irradiation-induced cell cycle checkpoints". Cancer Research. 57 (9): 1664–7. PMID 9135004.
- ^ Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE (septembar 2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. PMID 16169070.
- ^ Yan C, Wang H, Boyd DD (mart 2002). "ATF3 represses 72-kDa type IV collagenase (MMP-2) expression by antagonizing p53-dependent trans-activation of the collagenase promoter". The Journal of Biological Chemistry. 277 (13): 10804–12. doi:10.1074/jbc.M112069200. PMID 11792711.
- ^ Chen SS, Chang PC, Cheng YW, Tang FM, Lin YS (septembar 2002). "Suppression of the STK15 oncogenic activity requires a transactivation-independent p53 function". The EMBO Journal. 21 (17): 4491–9. doi:10.1093/emboj/cdf409. PMC 126178. PMID 12198151.
- ^ Leu JI, Dumont P, Hafey M, Murphy ME, George DL (maj 2004). "Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex". Nature Cell Biology. 6 (5): 443–50. doi:10.1038/ncb1123. PMID 15077116. S2CID 43063712.
- ^ a b c d e f Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R (novembar 2003). "Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair". Molecular Cell. 12 (5): 1087–99. doi:10.1016/S1097-2765(03)00424-6. PMID 14636569.
- ^ a b c Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, Halazonetis TD, Harris CC (septembar 2004). "Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest". The Journal of Cell Biology. 166 (6): 801–13. doi:10.1083/jcb.200405128. PMC 2172115. PMID 15364958.
- ^ Wang XW, Tseng A, Ellis NA, Spillare EA, Linke SP, Robles AI, Seker H, Yang Q, Hu P, Beresten S, Bemmels NA, Garfield S, Harris CC (august 2001). "Functional interaction of p53 and BLM DNA helicase in apoptosis". The Journal of Biological Chemistry. 276 (35): 32948–55. doi:10.1074/jbc.M103298200. PMID 11399766.
- ^ Garkavtsev IV, Kley N, Grigorian IA, Gudkov AV (decembar 2001). "The Bloom syndrome protein interacts and cooperates with p53 in regulation of transcription and cell growth control". Oncogene. 20 (57): 8276–80. doi:10.1038/sj.onc.1205120. PMID 11781842.
- ^ a b Yang Q, Zhang R, Wang XW, Spillare EA, Linke SP, Subramanian D, Griffith JD, Li JL, Hickson ID, Shen JC, Loeb LA, Mazur SJ, Appella E, Brosh RM, Karmakar P, Bohr VA, Harris CC (august 2002). "The processing of Holliday junctions by BLM and WRN helicases is regulated by p53". The Journal of Biological Chemistry. 277 (35): 31980–7. doi:10.1074/jbc.M204111200. PMID 12080066.
- ^ Abramovitch S, Werner H (2003). "Functional and physical interactions between BRCA1 and p53 in transcriptional regulation of the IGF-IR gene". Hormone and Metabolic Research. 35 (11–12): 758–62. doi:10.1055/s-2004-814154. PMID 14710355.
- ^ Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H (mart 1998). "BRCA1 regulates p53-dependent gene expression". Proceedings of the National Academy of Sciences of the United States of America. 95 (5): 2302–6. Bibcode:1998PNAS...95.2302O. doi:10.1073/pnas.95.5.2302. PMC 19327. PMID 9482880.
- ^ Chai YL, Cui J, Shao N, Shyam E, Reddy P, Rao VN (januar 1999). "The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter". Oncogene. 18 (1): 263–8. doi:10.1038/sj.onc.1202323. PMID 9926942.
- ^ Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, Weber BL, El-Deiry WS (april 1998). "BRCA1 physically associates with p53 and stimulates its transcriptional activity". Oncogene. 16 (13): 1713–21. doi:10.1038/sj.onc.1201932. PMID 9582019.
- ^ Marmorstein LY, Ouchi T, Aaronson SA (novembar 1998). "The BRCA2 gene product functionally interacts with p53 and RAD51". Proceedings of the National Academy of Sciences of the United States of America. 95 (23): 13869–74. Bibcode:1998PNAS...9513869M. doi:10.1073/pnas.95.23.13869. PMC 24938. PMID 9811893.
- ^ Uramoto H, Izumi H, Nagatani G, Ohmori H, Nagasue N, Ise T, Yoshida T, Yasumoto K, Kohno K (april 2003). "Physical interaction of tumour suppressor p53/p73 with CCAAT-binding transcription factor 2 (CTF2) and differential regulation of human high-mobility group 1 (HMG1) gene expression". The Biochemical Journal. 371 (Pt 2): 301–10. doi:10.1042/BJ20021646. PMC 1223307. PMID 12534345.
- ^ a b Li L, Ljungman M, Dixon JE (januar 2000). "The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53". The Journal of Biological Chemistry. 275 (4): 2410–4. doi:10.1074/jbc.275.4.2410. PMID 10644693.
- ^ Luciani MG, Hutchins JR, Zheleva D, Hupp TR (juli 2000). "The C-terminal regulatory domain of p53 contains a functional docking site for cyclin A". Journal of Molecular Biology. 300 (3): 503–18. doi:10.1006/jmbi.2000.3830. PMID 10884347.
- ^ Ababneh M, Götz C, Montenarh M (maj 2001). "Downregulation of the cdc2/cyclin B protein kinase activity by binding of p53 to p34(cdc2)". Biochemical and Biophysical Research Communications. 283 (2): 507–12. doi:10.1006/bbrc.2001.4792. PMID 11327730.
- ^ Abedini MR, Muller EJ, Brun J, Bergeron R, Gray DA, Tsang BK (juni 2008). "Cisplatin induces p53-dependent FLICE-like inhibitory protein ubiquitination in ovarian cancer cells". Cancer Research. 68 (12): 4511–7. doi:10.1158/0008-5472.CAN-08-0673. PMID 18559494.
- ^ a b Goudelock DM, Jiang K, Pereira E, Russell B, Sanchez Y (august 2003). "Regulatory interactions between the checkpoint kinase Chk1 and the proteins of the DNA-dependent protein kinase complex". The Journal of Biological Chemistry. 278 (32): 29940–7. doi:10.1074/jbc.M301765200. PMID 12756247.
- ^ Tian H, Faje AT, Lee SL, Jorgensen TJ (2002). "Radiation-induced phosphorylation of Chk1 at S345 is associated with p53-dependent cell cycle arrest pathways". Neoplasia. 4 (2): 171–80. doi:10.1038/sj.neo.7900219. PMC 1550321. PMID 11896572.
- ^ Zhao L, Samuels T, Winckler S, Korgaonkar C, Tompkins V, Horne MC, Quelle DE (januar 2003). "Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways". Molecular Cancer Research. 1 (3): 195–206. PMID 12556559.
- ^ a b Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP (novembar 2002). "MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation". The EMBO Journal. 21 (22): 6236–45. doi:10.1093/emboj/cdf616. PMC 137207. PMID 12426395.
- ^ a b Livengood JA, Scoggin KE, Van Orden K, McBryant SJ, Edayathumangalam RS, Laybourn PJ, Nyborg JK (mart 2002). "p53 Transcriptional activity is mediated through the SRC1-interacting domain of CBP/p300". The Journal of Biological Chemistry. 277 (11): 9054–61. doi:10.1074/jbc.M108870200. PMID 11782467.
- ^ a b Giebler HA, Lemasson I, Nyborg JK (juli 2000). "p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation". Molecular and Cellular Biology. 20 (13): 4849–58. doi:10.1128/MCB.20.13.4849-4858.2000. PMC 85936. PMID 10848610.
- ^ a b Schneider E, Montenarh M, Wagner P (novembar 1998). "Regulation of CAK kinase activity by p53". Oncogene. 17 (21): 2733–41. doi:10.1038/sj.onc.1202504. PMID 9840937.
- ^ a b Ko LJ, Shieh SY, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan ZQ (decembar 1997). "p53 is phosphorylated by CDK7-cyclin H in a p36MAT1-dependent manner". Molecular and Cellular Biology. 17 (12): 7220–9. doi:10.1128/mcb.17.12.7220. PMC 232579. PMID 9372954.
- ^ Yavuzer U, Smith GC, Bliss T, Werner D, Jackson SP (juli 1998). "DNA end-independent activation of DNA-PK mediated via association with the DNA-binding protein C1D". Genes & Development. 12 (14): 2188–99. doi:10.1101/gad.12.14.2188. PMC 317006. PMID 9679063.
- ^ a b Rizos H, Diefenbach E, Badhwar P, Woodruff S, Becker TM, Rooney RJ, Kefford RF (februar 2003). "Association of p14ARF with the p120E4F transcriptional repressor enhances cell cycle inhibition". The Journal of Biological Chemistry. 278 (7): 4981–9. doi:10.1074/jbc.M210978200. PMID 12446718.
- ^ Sandy P, Gostissa M, Fogal V, Cecco LD, Szalay K, Rooney RJ, Schneider C, Del Sal G (januar 2000). "p53 is involved in the p120E4F-mediated growth arrest". Oncogene. 19 (2): 188–99. doi:10.1038/sj.onc.1203250. PMID 10644996.
- ^ a b c Gallagher WM, Argentini M, Sierra V, Bracco L, Debussche L, Conseiller E (juni 1999). "MBP1: a novel mutant p53-specific protein partner with oncogenic properties". Oncogene. 18 (24): 3608–16. doi:10.1038/sj.onc.1202937. PMID 10380882.
- ^ Cuddihy AR, Wong AH, Tam NW, Li S, Koromilas AE (april 1999). "The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro". Oncogene. 18 (17): 2690–702. doi:10.1038/sj.onc.1202620. PMID 10348343.
- ^ Shinobu N, Maeda T, Aso T, Ito T, Kondo T, Koike K, Hatakeyama M (juni 1999). "Physical interaction and functional antagonism between the RNA polymerase II elongation factor ELL and p53". The Journal of Biological Chemistry. 274 (24): 17003–10. doi:10.1074/jbc.274.24.17003. PMID 10358050.
- ^ Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, Howley PM, Livingston DM (oktobar 1998). "p300/MDM2 complexes participate in MDM2-mediated p53 degradation". Molecular Cell. 2 (4): 405–15. doi:10.1016/S1097-2765(00)80140-9. PMID 9809062.
- ^ An W, Kim J, Roeder RG (juni 2004). "Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53". Cell. 117 (6): 735–48. doi:10.1016/j.cell.2004.05.009. PMID 15186775.
- ^ Pastorcic M, Das HK (novembar 2000). "Regulation of transcription of the human presenilin-1 gene by ets transcription factors and the p53 protooncogene". The Journal of Biological Chemistry. 275 (45): 34938–45. doi:10.1074/jbc.M005411200. PMID 10942770.
- ^ a b Wang XW, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly JM, Wang Z, Freidberg EC, Evans MK, Taffe BG (juni 1995). "p53 modulation of TFIIH-associated nucleotide excision repair activity". Nature Genetics. 10 (2): 188–95. doi:10.1038/ng0695-188. hdl:1765/54884. PMID 7663514. S2CID 38325851.
- ^ Yu A, Fan HY, Liao D, Bailey AD, Weiner AM (maj 2000). "Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes". Molecular Cell. 5 (5): 801–10. doi:10.1016/S1097-2765(00)80320-2. PMID 10882116.
- ^ Tsai RY, McKay RD (decembar 2002). "A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells". Genes & Development. 16 (23): 2991–3003. doi:10.1101/gad.55671. PMC 187487. PMID 12464630.
- ^ Peng YC, Kuo F, Breiding DE, Wang YF, Mansur CP, Androphy EJ (septembar 2001). "AMF1 (GPS2) modulates p53 transactivation". Molecular and Cellular Biology. 21 (17): 5913–24. doi:10.1128/MCB.21.17.5913-5924.2001. PMC 87310. PMID 11486030.
- ^ Watcharasit P, Bijur GN, Zmijewski JW, Song L, Zmijewska A, Chen X, Johnson GV, Jope RS (juni 2002). "Direct, activating interaction between glycogen synthase kinase-3beta and p53 after DNA damage". Proceedings of the National Academy of Sciences of the United States of America. 99 (12): 7951–5. Bibcode:2002PNAS...99.7951W. doi:10.1073/pnas.122062299. PMC 123001. PMID 12048243.
- ^ a b Akakura S, Yoshida M, Yoneda Y, Horinouchi S (maj 2001). "A role for Hsc70 in regulating nucleocytoplasmic transport of a temperature-sensitive p53 (p53Val-135)". The Journal of Biological Chemistry. 276 (18): 14649–57. doi:10.1074/jbc.M100200200. PMID 11297531.
- ^ Wang C, Chen J (januar 2003). "Phosphorylation and hsp90 binding mediate heat shock stabilization of p53". The Journal of Biological Chemistry. 278 (3): 2066–71. doi:10.1074/jbc.M206697200. PMID 12427754.
- ^ Peng Y, Chen L, Li C, Lu W, Chen J (novembar 2001). "Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization". The Journal of Biological Chemistry. 276 (44): 40583–90. doi:10.1074/jbc.M102817200. PMID 11507088.
- ^ Chen D, Li M, Luo J, Gu W (april 2003). "Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function". The Journal of Biological Chemistry. 278 (16): 13595–8. doi:10.1074/jbc.C200694200. PMID 12606552.
- ^ Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A (januar 2000). "Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha". Genes & Development. 14 (1): 34–44. doi:10.1101/gad.14.1.34 (neaktivno 31. 5. 2021). PMC 316350. PMID 10640274.CS1 održavanje: DOI nije aktivan od 2021 (link)
- ^ Hansson LO, Friedler A, Freund S, Rudiger S, Fersht AR (august 2002). "Two sequence motifs from HIF-1alpha bind to the DNA-binding site of p53". Proceedings of the National Academy of Sciences of the United States of America. 99 (16): 10305–9. Bibcode:2002PNAS...9910305H. doi:10.1073/pnas.122347199. PMC 124909. PMID 12124396.
- ^ An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM (mart 1998). "Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha". Nature. 392 (6674): 405–8. Bibcode:1998Natur.392..405A. doi:10.1038/32925. PMID 9537326. S2CID 4423081.
- ^ Kondo S, Lu Y, Debbas M, Lin AW, Sarosi I, Itie A, Wakeham A, Tuan J, Saris C, Elliott G, Ma W, Benchimol S, Lowe SW, Mak TW, Thukral SK (april 2003). "Characterization of cells and gene-targeted mice deficient for the p53-binding kinase homeodomain-interacting protein kinase 1 (HIPK1)". Proceedings of the National Academy of Sciences of the United States of America. 100 (9): 5431–6. Bibcode:2003PNAS..100.5431K. doi:10.1073/pnas.0530308100. PMC 154362. PMID 12702766.
- ^ Hofmann TG, Möller A, Sirma H, Zentgraf H, Taya Y, Dröge W, Will H, Schmitz ML (januar 2002). "Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2". Nature Cell Biology. 4 (1): 1–10. doi:10.1038/ncb715. PMID 11740489. S2CID 37789883.
- ^ Kim EJ, Park JS, Um SJ (august 2002). "Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo". The Journal of Biological Chemistry. 277 (35): 32020–8. doi:10.1074/jbc.M200153200. PMID 11925430.
- ^ Imamura T, Izumi H, Nagatani G, Ise T, Nomoto M, Iwamoto Y, Kohno K (mart 2001). "Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein". The Journal of Biological Chemistry. 276 (10): 7534–40. doi:10.1074/jbc.M008143200. PMID 11106654.
- ^ Dintilhac A, Bernués J (mart 2002). "HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences". The Journal of Biological Chemistry. 277 (9): 7021–8. doi:10.1074/jbc.M108417200. PMID 11748221.
- ^ Wadhwa R, Yaguchi T, Hasan MK, Mitsui Y, Reddel RR, Kaul SC (april 2002). "Hsp70 family member, mot-2/mthsp70/GRP75, binds to the cytoplasmic sequestration domain of the p53 protein". Experimental Cell Research. 274 (2): 246–53. doi:10.1006/excr.2002.5468. PMID 11900485.
- ^ Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM (juni 2000). "The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription". Proceedings of the National Academy of Sciences of the United States of America. 97 (12): 6763–8. Bibcode:2000PNAS...97.6763S. doi:10.1073/pnas.100110097. PMC 18731. PMID 10823891.
- ^ Leung KM, Po LS, Tsang FC, Siu WY, Lau A, Ho HT, Poon RY (septembar 2002). "The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2". Cancer Research. 62 (17): 4890–3. PMID 12208736.
- ^ Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV (januar 1998). "The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control". Nature. 391 (6664): 295–8. Bibcode:1998Natur.391..295G. doi:10.1038/34675. PMID 9440695. S2CID 4429461.
- ^ a b Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC (maj 2003). "p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity". Cancer Research. 63 (10): 2373–8. PMID 12750254.
- ^ Tsai KW, Tseng HC, Lin WC (oktobar 2008). "Two wobble-splicing events affect ING4 protein subnuclear localization and degradation". Experimental Cell Research. 314 (17): 3130–41. doi:10.1016/j.yexcr.2008.08.002. PMID 18775696.
- ^ Chang NS (mart 2002). "The non-ankyrin C terminus of Ikappa Balpha physically interacts with p53 in vivo and dissociates in response to apoptotic stress, hypoxia, DNA damage, and transforming growth factor-beta 1-mediated growth suppression". The Journal of Biological Chemistry. 277 (12): 10323–31. doi:10.1074/jbc.M106607200. PMID 11799106.
- ^ a b Kurki S, Latonen L, Laiho M (oktobar 2003). "Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization". Journal of Cell Science. 116 (Pt 19): 3917–25. doi:10.1242/jcs.00714. PMID 12915590.
- ^ a b Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, Liu X, Wu H (februar 2003). "PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms". Cancer Cell. 3 (2): 117–30. doi:10.1016/S1535-6108(03)00021-7. PMID 12620407.
- ^ a b Zhang Y, Xiong Y, Yarbrough WG (mart 1998). "ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways". Cell. 92 (6): 725–34. doi:10.1016/S0092-8674(00)81401-4. PMID 9529249.
- ^ Badciong JC, Haas AL (decembar 2002). "MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination". The Journal of Biological Chemistry. 277 (51): 49668–75. doi:10.1074/jbc.M208593200. PMID 12393902.
- ^ Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G, van Ham RC, van der Houven van Oordt W, van der Eb AJ, Jochemsen AG (juli 1997). "Isolation and identification of the human homolog of a new p53-binding protein, Mdmx" (PDF). Genomics. 43 (1): 34–42. doi:10.1006/geno.1997.4775. hdl:2066/142231. PMID 9226370.
- ^ Frade R, Balbo M, Barel M (decembar 2000). "RB18A, whose gene is localized on chromosome 17q12-q21.1, regulates in vivo p53 transactivating activity". Cancer Research. 60 (23): 6585–9. PMID 11118038.
- ^ Drané P, Barel M, Balbo M, Frade R (decembar 1997). "Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53". Oncogene. 15 (25): 3013–24. doi:10.1038/sj.onc.1201492. PMID 9444950.
- ^ Hu MC, Qiu WR, Wang YP (novembar 1997). "JNK1, JNK2 and JNK3 are p53 N-terminal serine 34 kinases". Oncogene. 15 (19): 2277–87. doi:10.1038/sj.onc.1201401. PMID 9393873.
- ^ Lin Y, Khokhlatchev A, Figeys D, Avruch J (decembar 2002). "Death-associated protein 4 binds MST1 and augments MST1-induced apoptosis". The Journal of Biological Chemistry. 277 (50): 47991–8001. doi:10.1074/jbc.M202630200. PMID 12384512.
- ^ Taniura H, Matsumoto K, Yoshikawa K (juni 1999). "Physical and functional interactions of neuronal growth suppressor necdin with p53". The Journal of Biological Chemistry. 274 (23): 16242–8. doi:10.1074/jbc.274.23.16242. PMID 10347180.
- ^ Daniely Y, Dimitrova DD, Borowiec JA (august 2002). "Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation". Molecular and Cellular Biology. 22 (16): 6014–22. doi:10.1128/MCB.22.16.6014-6022.2002. PMC 133981. PMID 12138209.
- ^ Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP (januar 2008). "NUMB controls p53 tumour suppressor activity". Nature. 451 (7174): 76–80. Bibcode:2008Natur.451...76C. doi:10.1038/nature06412. PMID 18172499. S2CID 4431258.
- ^ a b c Choy MK, Movassagh M, Siggens L, Vujic A, Goddard M, Sánchez A, Perkins N, Figg N, Bennett M, Carroll J, Foo R (juni 2010). "High-throughput sequencing identifies STAT3 as the DNA-associated factor for p53-NF-kappaB-complex-dependent gene expression in human heart failure". Genome Medicine. 2 (6): 37. doi:10.1186/gm158. PMC 2905097. PMID 20546595.
- ^ a b Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y (decembar 2003). "Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway". Molecular and Cellular Biology. 23 (23): 8902–12. doi:10.1128/MCB.23.23.8902-8912.2003. PMC 262682. PMID 14612427.
- ^ Nikolaev AY, Li M, Puskas N, Qin J, Gu W (januar 2003). "Parc: a cytoplasmic anchor for p53". Cell. 112 (1): 29–40. doi:10.1016/S0092-8674(02)01255-2. PMID 12526791.
- ^ Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR (maj 1998). "Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions". The Journal of Biological Chemistry. 273 (19): 11839–43. doi:10.1074/jbc.273.19.11839. PMID 9565608.
- ^ Kahyo T, Nishida T, Yasuda H (septembar 2001). "Involvement of PIAS1 in the sumoylation of tumor suppressor p53". Molecular Cell. 8 (3): 713–8. doi:10.1016/S1097-2765(01)00349-5. PMID 11583632.
- ^ Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP (decembar 2002). "Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage". The Journal of Biological Chemistry. 277 (50): 47976–9. doi:10.1074/jbc.C200538200. PMID 12388558.
- ^ Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G (oktobar 2002). "The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults". Nature. 419 (6909): 853–7. Bibcode:2002Natur.419..853Z. doi:10.1038/nature01120. PMID 12397362. S2CID 4311658.
- ^ Huang SM, Schönthal AH, Stallcup MR (april 2001). "Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1". Oncogene. 20 (17): 2134–43. doi:10.1038/sj.onc.1204298. PMID 11360197.
- ^ Xie S, Wu H, Wang Q, Cogswell JP, Husain I, Conn C, Stambrook P, Jhanwar-Uniyal M, Dai W (novembar 2001). "Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway". The Journal of Biological Chemistry. 276 (46): 43305–12. doi:10.1074/jbc.M106050200. PMID 11551930.
- ^ Bahassi EM, Conn CW, Myer DL, Hennigan RF, McGowan CH, Sanchez Y, Stambrook PJ (septembar 2002). "Mammalian Polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways". Oncogene. 21 (43): 6633–40. doi:10.1038/sj.onc.1205850. PMID 12242661.
- ^ Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V (januar 1997). "PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb". Oncogene. 14 (2): 145–55. doi:10.1038/sj.onc.1200825. PMID 9010216.
- ^ Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S (novembar 2003). "Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling". The Journal of Biological Chemistry. 278 (48): 47853–61. doi:10.1074/jbc.M305171200. PMID 14500729.
- ^ Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G (novembar 2000). "Regulation of p53 activity in nuclear bodies by a specific PML isoform". The EMBO Journal. 19 (22): 6185–95. doi:10.1093/emboj/19.22.6185. PMC 305840. PMID 11080164.
- ^ Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP (oktobar 2000). "The function of PML in p53-dependent apoptosis". Nature Cell Biology. 2 (10): 730–6. doi:10.1038/35036365. PMID 11025664. S2CID 13480833.
- ^ a b Zhang Z, Zhang R (mart 2008). "Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation". The EMBO Journal. 27 (6): 852–64. doi:10.1038/emboj.2008.25. PMC 2265109. PMID 18309296.
- ^ Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D (januar 2008). "Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation". Molecular Cell. 29 (1): 9–22. doi:10.1016/j.molcel.2007.11.031. PMC 2234035. PMID 18206965.
- ^ Bernal JA, Luna R, Espina A, Lázaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA (oktobar 2002). "Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis". Nature Genetics. 32 (2): 306–11. doi:10.1038/ng997. PMID 12355087. S2CID 1770399.
- ^ Stürzbecher HW, Donzelmann B, Henning W, Knippschild U, Buchhop S (april 1996). "p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction". The EMBO Journal. 15 (8): 1992–2002. doi:10.1002/j.1460-2075.1996.tb00550.x. PMC 450118. PMID 8617246.
- ^ Buchhop S, Gibson MK, Wang XW, Wagner P, Stürzbecher HW, Harris CC (oktobar 1997). "Interaction of p53 with the human Rad51 protein". Nucleic Acids Research. 25 (19): 3868–74. doi:10.1093/nar/25.19.3868. PMC 146972. PMID 9380510.
- ^ Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S (mart 2003). "Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation". Cell. 112 (6): 779–91. doi:10.1016/S0092-8674(03)00193-4. PMID 12654245.
- ^ Sheng Y, Laister RC, Lemak A, Wu B, Tai E, Duan S, Lukin J, Sunnerhagen M, Srisailam S, Karra M, Benchimol S, Arrowsmith CH (decembar 2008). "Molecular basis of Pirh2-mediated p53 ubiquitylation". Nature Structural & Molecular Biology. 15 (12): 1334–42. doi:10.1038/nsmb.1521. PMC 4075976. PMID 19043414.
- ^ Romanova LY, Willers H, Blagosklonny MV, Powell SN (decembar 2004). "The interaction of p53 with replication protein A mediates suppression of homologous recombination". Oncogene. 23 (56): 9025–33. doi:10.1038/sj.onc.1207982. PMID 15489903.
- ^ Riva F, Zuco V, Vink AA, Supino R, Prosperi E (decembar 2001). "UV-induced DNA incision and proliferating cell nuclear antigen recruitment to repair sites occur independently of p53-replication protein A interaction in p53 wild type and mutant ovarian carcinoma cells". Carcinogenesis. 22 (12): 1971–8. doi:10.1093/carcin/22.12.1971. PMID 11751427.
- ^ Lin J, Yang Q, Yan Z, Markowitz J, Wilder PT, Carrier F, Weber DJ (august 2004). "Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells". The Journal of Biological Chemistry. 279 (32): 34071–7. doi:10.1074/jbc.M405419200. PMID 15178678.
- ^ a b Minty A, Dumont X, Kaghad M, Caput D (novembar 2000). "Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif". The Journal of Biological Chemistry. 275 (46): 36316–23. doi:10.1074/jbc.M004293200. PMID 10961991.
- ^ a b Ivanchuk SM, Mondal S, Rutka JT (juni 2008). "p14ARF interacts with DAXX: effects on HDM2 and p53". Cell Cycle. 7 (12): 1836–50. doi:10.4161/cc.7.12.6025. PMID 18583933.
- ^ a b Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J (juni 2002). "SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription". The Journal of Biological Chemistry. 277 (25): 22330–7. doi:10.1074/jbc.M111987200. PMID 11950834.
- ^ Young PJ, Day PM, Zhou J, Androphy EJ, Morris GE, Lorson CL (januar 2002). "A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy". The Journal of Biological Chemistry. 277 (4): 2852–9. doi:10.1074/jbc.M108769200. PMID 11704667.
- ^ Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T (decembar 1992). "Wild-type p53 binds to the TATA-binding protein and represses transcription". Proceedings of the National Academy of Sciences of the United States of America. 89 (24): 12028–32. Bibcode:1992PNAS...8912028S. doi:10.1073/pnas.89.24.12028. PMC 50691. PMID 1465435.
- ^ Cvekl A, Kashanchi F, Brady JN, Piatigorsky J (juni 1999). "Pax-6 interactions with TATA-box-binding protein and retinoblastoma protein". Investigative Ophthalmology & Visual Science. 40 (7): 1343–50. PMID 10359315.
- ^ McPherson LA, Loktev AV, Weigel RJ (novembar 2002). "Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53". The Journal of Biological Chemistry. 277 (47): 45028–33. doi:10.1074/jbc.M208924200. PMID 12226108.
- ^ Sørensen TS, Girling R, Lee CW, Gannon J, Bandara LR, La Thangue NB (oktobar 1996). "Functional interaction between DP-1 and p53". Molecular and Cellular Biology. 16 (10): 5888–95. doi:10.1128/mcb.16.10.5888. PMC 231590. PMID 8816502.
- ^ Green DR, Chipuk JE (juli 2006). "p53 and metabolism: Inside the TIGAR". Cell. 126 (1): 30–2. doi:10.1016/j.cell.2006.06.032. PMID 16839873.
- ^ Gobert C, Skladanowski A, Larsen AK (august 1999). "The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53". Proceedings of the National Academy of Sciences of the United States of America. 96 (18): 10355–60. Bibcode:1999PNAS...9610355G. doi:10.1073/pnas.96.18.10355. PMC 17892. PMID 10468612.
- ^ Mao Y, Mehl IR, Muller MT (februar 2002). "Subnuclear distribution of topoisomerase I is linked to ongoing transcription and p53 status". Proceedings of the National Academy of Sciences of the United States of America. 99 (3): 1235–40. Bibcode:2002PNAS...99.1235M. doi:10.1073/pnas.022631899. PMC 122173. PMID 11805286.
- ^ a b Cowell IG, Okorokov AL, Cutts SA, Padget K, Bell M, Milner J, Austin CA (februar 2000). "Human topoisomerase IIalpha and IIbeta interact with the C-terminal region of p53". Experimental Cell Research. 255 (1): 86–94. doi:10.1006/excr.1999.4772. PMID 10666337.
- ^ Derbyshire DJ, Basu BP, Serpell LC, Joo WS, Date T, Iwabuchi K, Doherty AJ (juli 2002). "Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor". The EMBO Journal. 21 (14): 3863–72. doi:10.1093/emboj/cdf383. PMC 126127. PMID 12110597.
- ^ Ekblad CM, Friedler A, Veprintsev D, Weinberg RL, Itzhaki LS (mart 2004). "Comparison of BRCT domains of BRCA1 and 53BP1: a biophysical analysis". Protein Science. 13 (3): 617–25. doi:10.1110/ps.03461404. PMC 2286730. PMID 14978302.
- ^ Lo KW, Kan HM, Chan LN, Xu WG, Wang KP, Wu Z, Sheng M, Zhang M (mart 2005). "The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation". The Journal of Biological Chemistry. 280 (9): 8172–9. doi:10.1074/jbc.M411408200. PMID 15611139.
- ^ Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP (mart 2002). "Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure". Genes & Development. 16 (5): 583–93. doi:10.1101/gad.959202. PMC 155350. PMID 11877378.
- ^ Derbyshire DJ, Basu BP, Date T, Iwabuchi K, Doherty AJ (oktobar 2002). "Purification, crystallization and preliminary X-ray analysis of the BRCT domains of human 53BP1 bound to the p53 tumour suppressor". Acta Crystallographica D. 58 (Pt 10 Pt 2): 1826–9. doi:10.1107/S0907444902010910. PMID 12351827.
- ^ a b Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S (juni 1994). "Two cellular proteins that bind to wild-type but not mutant p53". Proceedings of the National Academy of Sciences of the United States of America. 91 (13): 6098–102. Bibcode:1994PNAS...91.6098I. doi:10.1073/pnas.91.13.6098. PMC 44145. PMID 8016121.
- ^ Naumovski L, Cleary ML (juli 1996). "The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M". Molecular and Cellular Biology. 16 (7): 3884–92. doi:10.1128/MCB.16.7.3884. PMC 231385. PMID 8668206.
- ^ Tomasini R, Samir AA, Carrier A, Isnardon D, Cecchinelli B, Soddu S, Malissen B, Dagorn JC, Iovanna JL, Dusetti NJ (septembar 2003). "TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity". The Journal of Biological Chemistry. 278 (39): 37722–9. doi:10.1074/jbc.M301979200. PMID 12851404.
- ^ Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y (juli 2001). "p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis". Molecular Cell. 8 (1): 85–94. doi:10.1016/S1097-2765(01)00284-2. PMID 11511362.
- ^ Li L, Liao J, Ruland J, Mak TW, Cohen SN (februar 2001). "A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control". Proceedings of the National Academy of Sciences of the United States of America. 98 (4): 1619–24. Bibcode:2001PNAS...98.1619L. doi:10.1073/pnas.98.4.1619. PMC 29306. PMID 11172000.
- ^ Lyakhovich A, Shekhar MP (april 2003). "Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response". Molecular and Cellular Biology. 23 (7): 2463–75. doi:10.1128/MCB.23.7.2463-2475.2003. PMC 150718. PMID 12640129.
- ^ Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ (oktobar 1996). "Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system". Genomics. 37 (2): 183–6. doi:10.1006/geno.1996.0540. PMID 8921390.
- ^ Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (februar 2002). "Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1". Cell. 108 (3): 345–56. doi:10.1016/S0092-8674(02)00630-X. PMID 11853669.
- ^ Sehat B, Andersson S, Girnita L, Larsson O (juli 2008). "Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis". Cancer Research. 68 (14): 5669–77. doi:10.1158/0008-5472.CAN-07-6364. PMID 18632619.
- ^ Song MS, Song SJ, Kim SY, Oh HJ, Lim DS (juli 2008). "The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex". The EMBO Journal. 27 (13): 1863–74. doi:10.1038/emboj.2008.115. PMC 2486425. PMID 18566590.
- ^ Yang W, Dicker DT, Chen J, El-Deiry WS (mart 2008). "CARPs enhance p53 turnover by degrading 14-3-3sigma and stabilizing MDM2". Cell Cycle. 7 (5): 670–82. doi:10.4161/cc.7.5.5701. PMID 18382127.
- ^ Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, Oren M, Tanaka N (mart 2008). "Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2". Proceedings of the National Academy of Sciences of the United States of America. 105 (12): 4838–43. Bibcode:2008PNAS..105.4838A. doi:10.1073/pnas.0712216105. PMC 2290789. PMID 18359851.
- ^ Dohmesen C, Koeppel M, Dobbelstein M (januar 2008). "Specific inhibition of Mdm2-mediated neddylation by Tip60". Cell Cycle. 7 (2): 222–31. doi:10.4161/cc.7.2.5185. PMID 18264029.
- ^ Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (april 2002). "Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization". Nature. 416 (6881): 648–53. Bibcode:2002Natur.416..648L. doi:10.1038/nature737. PMID 11923872. S2CID 4389394.
- ^ Brosh RM, Karmakar P, Sommers JA, Yang Q, Wang XW, Spillare EA, Harris CC, Bohr VA (septembar 2001). "p53 Modulates the exonuclease activity of Werner syndrome protein". The Journal of Biological Chemistry. 276 (37): 35093–102. doi:10.1074/jbc.M103332200. PMID 11427532.
- ^ Chang NS, Pratt N, Heath J, Schultz L, Sleve D, Carey GB, Zevotek N (februar 2001). "Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity". The Journal of Biological Chemistry. 276 (5): 3361–70. doi:10.1074/jbc.M007140200. PMID 11058590.
- ^ Okamoto T, Izumi H, Imamura T, Takano H, Ise T, Uchiumi T, Kuwano M, Kohno K (decembar 2000). "Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression". Oncogene. 19 (54): 6194–202. doi:10.1038/sj.onc.1204029. PMID 11175333.
- ^ Kelley KD, Miller KR, Todd A, Kelley AR, Tuttle R, Berberich SJ (maj 2010). "YPEL3, a p53-regulated gene that induces cellular senescence". Cancer Research. 70 (9): 3566–75. doi:10.1158/0008-5472.CAN-09-3219. PMC 2862112. PMID 20388804.
- ^ Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD (juni 1998). "ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins". Nature Genetics. 19 (2): 175–8. doi:10.1038/542. PMID 9620776. S2CID 26600934.
- ^ Liu J, Grogan L, Nau MM, Allegra CJ, Chu E, Wright JJ (april 2001). "Physical interaction between p53 and primary response gene Egr-1". International Journal of Oncology. 18 (4): 863–70. doi:10.3892/ijo.18.4.863. PMID 11251186.
- ^ Bai L, Merchant JL (juli 2001). "ZBP-89 promotes growth arrest through stabilization of p53". Molecular and Cellular Biology. 21 (14): 4670–83. doi:10.1128/MCB.21.14.4670-4683.2001. PMC 87140. PMID 11416144.
- ^ Yamakuchi M, Lowenstein CJ (mart 2009). "MiR-34, SIRT1 and p53: the feedback loop". Cell Cycle. 8 (5): 712–5. doi:10.4161/cc.8.5.7753. PMID 19221490.
- ^ Wang Y, Zhang J, Li J, Gui R, Nie X, Huang R. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn J of Basic Med Sci [Internet]. 2019May20 [cited 2020Apr.7];19(2):155-63. Available from: https://www.bjbms.org/ojs/index.php/bjbms/article/view/3935
Vanjski linkovi
uredi- "p53 Knowledgebase". Lane Group at the Institute of Molecular and Cell Biology (IMCB), Singapore. Arhivirano s originala, 3. 1. 2006. Pristupljeno 6. 4. 2008.
- GeneReviews/NCBI/NIH/UW entry on Li-Fraumeni Syndrome
- TUMOR PROTEIN p53 @ OMIM
- p53 restoration of function
- p53 @ The Atlas of Genetics and Cytogenetics in Oncology and Haematology
- TP53 Gene @ GeneCards
- p53 News provided by insciences organisation
- David S. Goodsel l (1. 7. 2002). "p53 Tumor Suppressor". Molecule of the Month. RCSB Protein Data Bank. Pristupljeno 6. 4. 2008.
- Thierry Soussi. "p53 Web Site". Pristupljeno 6. 4. 2008.
- Living LFS A non-profit Li-Fraumeni Syndrome patient support organization
- The George Pantziarka TP53 Trust A support group from the UK for sufferers of Li-Fraumeni Syndrome or other TP53-related disorders
- IARC TP53 Somatic Mutations database Arhivirano 15. 8. 2012. na Wayback Machine maintained at IARC, Lyon, by Magali Olivier
- PDBe-KB provides an overview of all the structure information available in the PDB for Human P53.
- scientific animation conformational changes of p53 upon binding to DNA